Are Your Cells Changing During a Trypsinization Protocol for Cell Dissociation?
by Erica D'AngeloResearchers who regularly perform cell culture are likely familiar with the process of trypsinization. A trypsinization protocol is used for cell dissociation, in order to move cells from one environment to another. Usually cell dissociation is performed so that cells can be moved from the plate or flask in which they are grown to a new suspended medium, from which they can be separated and studied further. In this process, a trypsin EDTA solution is applied to the cells. Trypsin EDTA solutions contain both the trypsin enzyme, which breaks down proteins on the cell surface in order to dissociate them from the container and each other, and EDTA, which allows trypsin to break down specific bonds in order to achieve this cell dissociation. The trypsin enzyme is a digestive enzyme found in mammals. When using a trypsin enzyme in a trypsin EDTA solution, however, “cell surface proteins are often cleaved, which leads to dysregulation of the cell functions.”1
Another issue with trypsinization can be the time lag between taking cells out of the incubator and the addition of lysis buffer for downstream protein and RNA studies. This time window is usually around 15 to 20 minutes, during which changes in gene expression can happen. Changes in expression are possible even after only a few minutes and have already been described for some genes.2,3,4 These changes can have implications for downstream analysis and interpretation of RNA expression.
With a special protocol for CellCover this time gap is closed, which allows for the analysis of all proteins, RNA, DNA, and cell morphology at time point zero – the time at which cell samples are taken out of the incubator. With CellCover, the start of the trypsinization protocol and the time at which downstream analyses such as RNA sequencing happen are virtually indistinguishable.
CellCover exerts its stabilizing effect very fast, preventing cell degradation. DNA, RNA, protein stay in place in a close to native condition, without crosslinking activity. Harsh treatments with alcohol, acetone or formaldehyde can be avoided.
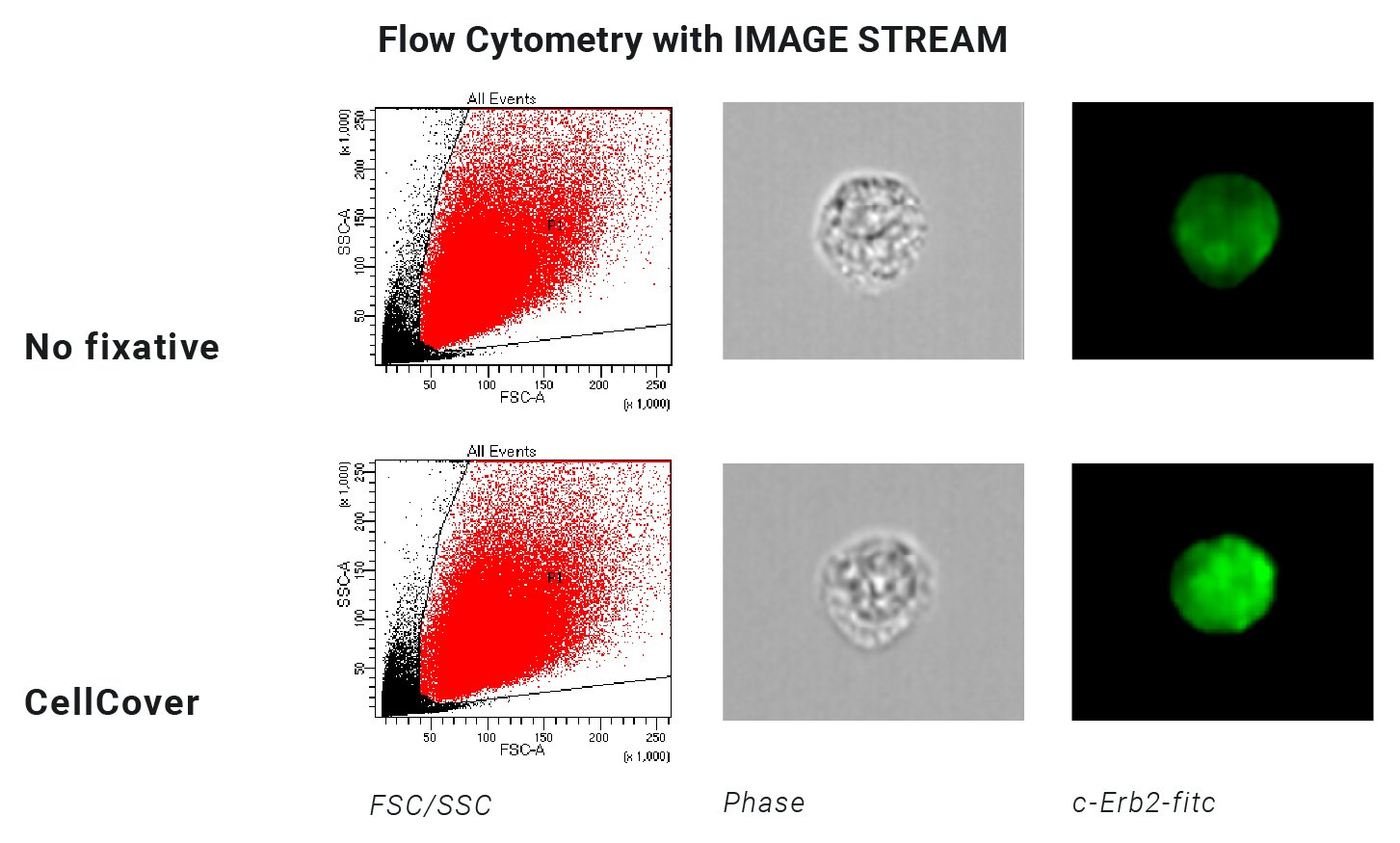
CellCover is the only reagent in the market which allows the parallel storage of proteins, RNA and DNA within it’s cellular context while maintaining the integrity of cellular shape without chemical crosslinking. This means for example, that you can take a heterogeneous cell population, discriminate different cells types by antigen mining (immunolabelling), sort them by flow cytometry and then isolate RNA, DNA or proteins for further analyses.
1Huang H.-L., Hsing H.-W., Lai T.-C., Chen Y.-W., Lee T.-R., Chan H.-T., Lyu P.-C., Wu C.-L., Lu Y.-C., Lin S.-T., et al. Trypsin-induced proteome alteration during cell subculture in mammalian cells. J. Biomed. Sci. 2010;17:36. doi: 10.1186/1423-0127-17-36.
2Thiel, C.S., Hauschild, S., Huge, A. et al. Dynamic gene expression response to altered gravity in human T cells. Sci Rep 7, 5204 (2017). https://doi.org/10.1038/s41598-017-05580-x
3Miyamoto S, Qin J, Safer B. Detection of early gene expression changes during activation of human primary lymphocytes by in vitro synthesis of proteins from polysomeassociated mRNAs. Protein Sci. 2001;10:423–433.
4Blanchard JL, Wholey W-Y, Conlon EM, Pomposiello PJ. 2007. Rapid changes in gene expression dynamics in response to superoxide reveal SoxRS-dependent and independent transcriptional networks. PLoS One 2:e1186. doi:10.1371/journal.pone.0001186.