verspark is a super-resolution microscopy mounting buffer, allowing you to image blinking dyes-labelled samples for several weeks. Everspark is compatible with multiple super-resolution microscopy techniques such as dSTORM, PALM, MINFLUX, or TIRF.
Sizes: Everspark 1.0: 4.5mL (10*450 µL) (KMO-ETE-450)
Everspark 2.0: 4.5mL (10*450 µL) KMO-ETE+-450-10
Catalogue Numbers: Everspark 1.0: KMO-ETE-450 (4.5mL (10*450 µL))
Everspark 2.0: KMO-ETE+-450-10 (4.5mL (10*450 µL))
Discover the long-lasting Everspark blinking buffer series for prolonged blinking in dSTORM microscopy. No more rushing your imaging. Mount your sample, and get stable blinking for weeks.
NEW ! Discover the newly-developed Everspark 2.0 also compatible with green fluorophores (i.e. AF488) for improved resolution in 3-colour dSTORM microscopy.- 10 vials with 450 µL of Everspark buffer at 100 mM MEA in Tris.
- 1 vial per experiment.
Vials lifetime: Up to 6 months in a closed pouch (e.g. unmounted).
Laser requirements: for green fluorophore imaging (Everspark 2.0 only), we recommend using a 488 laser >200mW, ideally 500mW for optimal results.
Additional information:
Product overviewSafety Datasheet
Certificate of Analysis
Results:
Co-localisation studies using three-colour 3D dSTORM imaging with Everspark 2.0 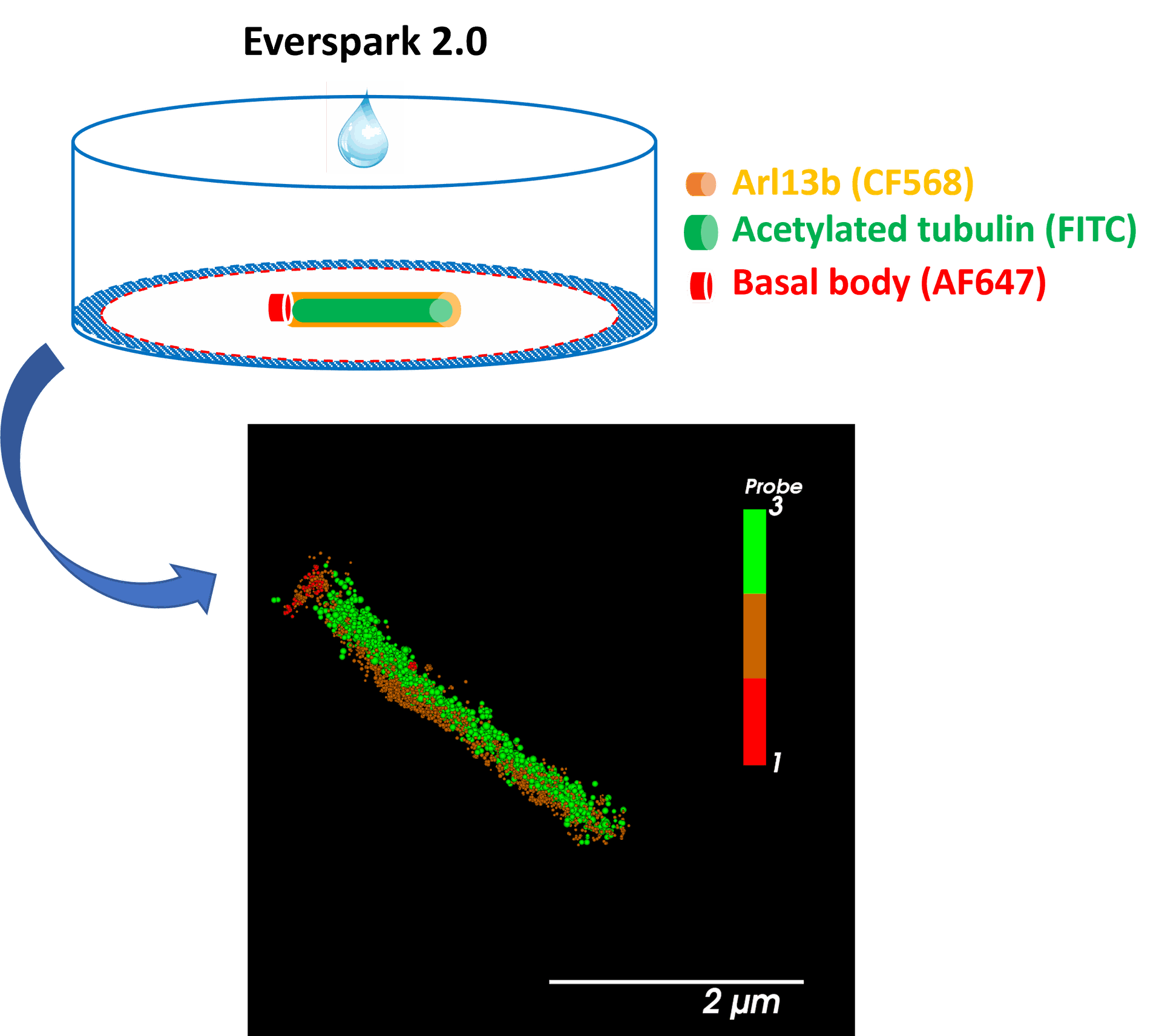 Centrosomal proteins Cep152, rootletin and PCM labelled with AF647, CF568 & AF488 respectively were imaged on a Vutara VXL (Bruker) inEverspark 2.0 buffer before 3D dSTORM reconstruction.
Centrosomal proteins Cep152, rootletin and PCM labelled with AF647, CF568 & AF488 respectively were imaged on a Vutara VXL (Bruker) inEverspark 2.0 buffer before 3D dSTORM reconstruction.
Credits: Karine Monier, INMG-PGNM, Lyon
Co-localisation studies using three-colour 3D dSTORM imaging with Everspark 2.0
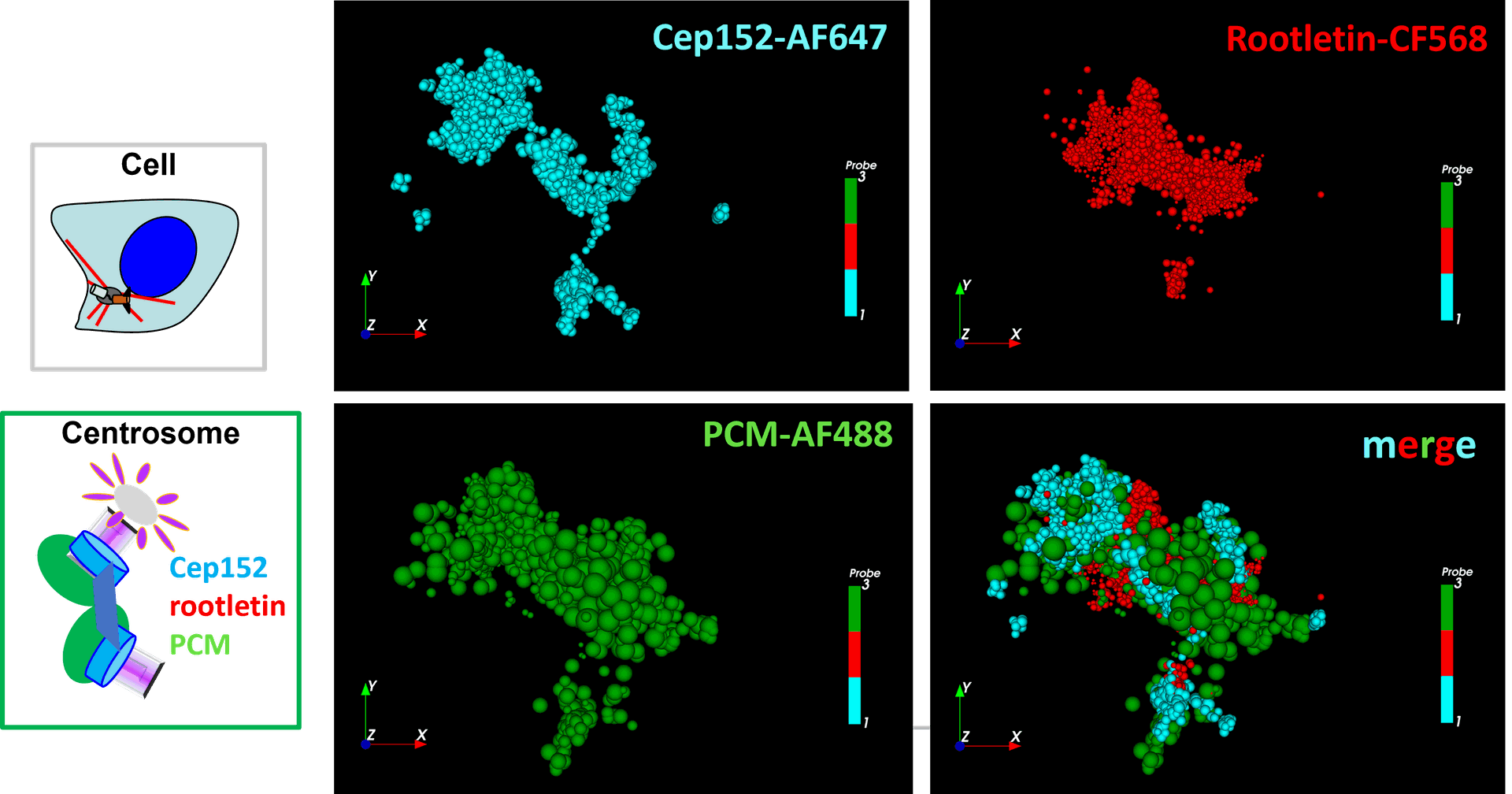
Centrosomal proteins Cep152, rootletin and PCM labelled with AF647, CF568 & AF488 respectively were imaged on a Vutara VXL (Bruker) in Everspark 2.0 buffer before 3D dSTORM reconstruction
Credits: Karine Monier, INMG-PGNM, Lyon
Green-channel dSTORM imaging of AF488 fluorophore using Everspark 2.0 buffer
AF488-coated beads were mounted in Everspark 1.0 or Everspark 2.0 buffer and imaged on a Vutara VXL (Bruker) before 3D dSTORM reconstruction.
Credits: Karine Monier, INMG-PGNM, Lyon
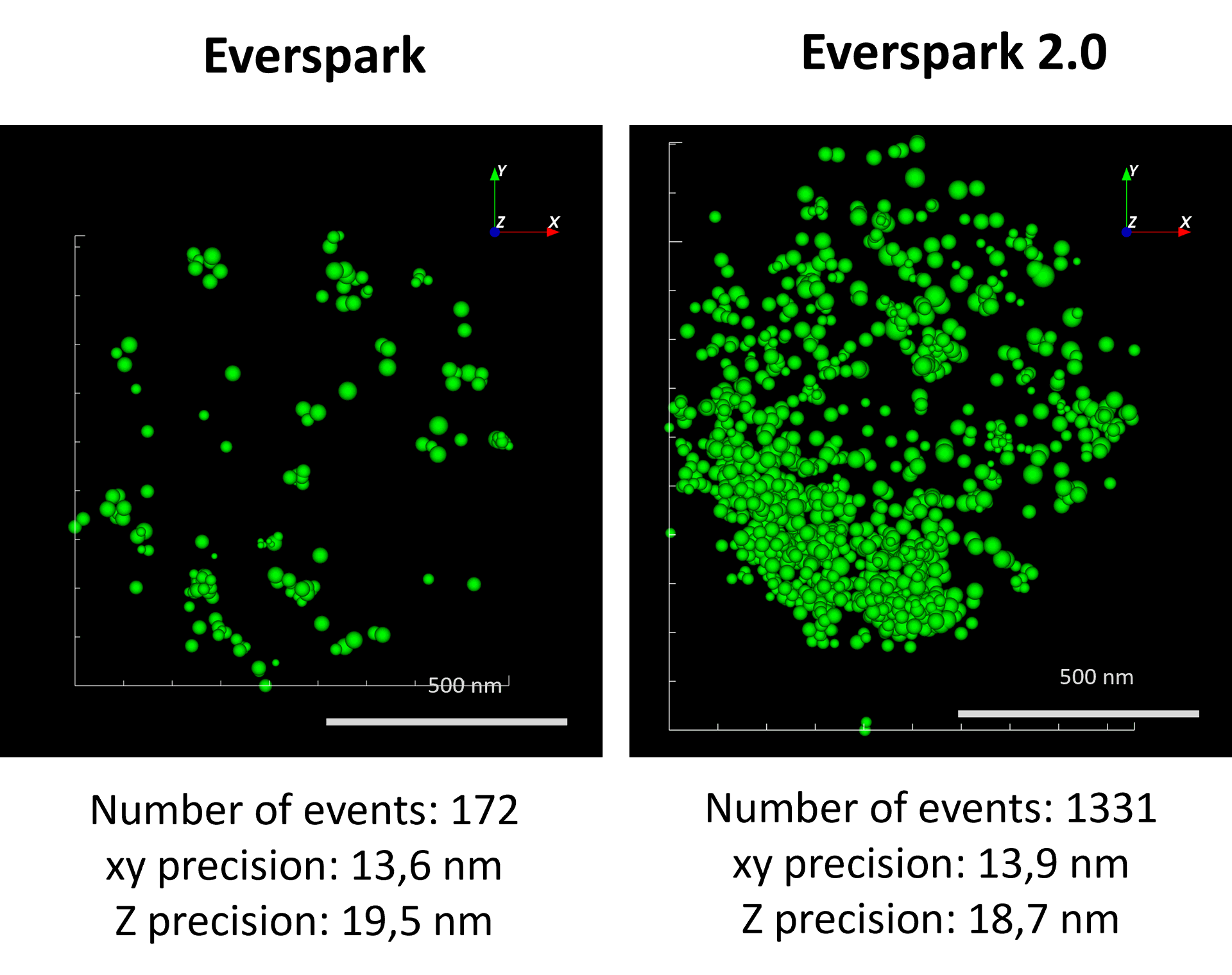
Long-term imaging with Everspark 1.0
Left: Two centrosomes imaged the same Day (D0) and 7 days after mounting (D7) on the same slide stored in the dark at 4°C. The typical 450 nm donut-like structure is visualised using a colour-coded scale encoded with the IGOR software, where each point appears as a function of its localisation precision (5 to 60 nm; inverted rainbow colour scale). Labeling: Distal-appendages detected by immunofluorescence with AF647 in RPE-1 cells.
Right: The number of blinking events per centrosome and the median of the localization precision in nm are presented for each serie of 50,000 images recorded at D0 and D7 (left).
Credits: Camille Fourneaux & Karine Monier, CRCL, Lyon
Donut-like structure of in-cellulo mature centrosome reconstructed after dSTORM imaging in Everspark 1.0 buffer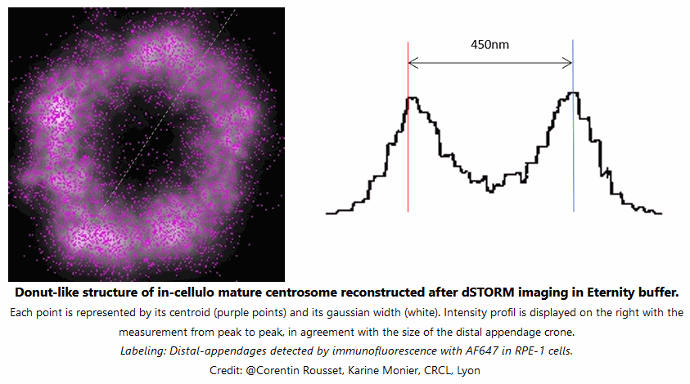
Each point is represented by its centroid (purple points) and its gaussian width (white). Intensity profile is displayed on the right with the measurement from peak to peak, in agreement with the size of the distal appendage crone. Labeling: distal-appendages detected by immunofluorescence with AF647 in RPE-1 cells.
Credits: Corentin Rousset, Karine Monier, CRCL Lyon





